Data Preprocessing and Extraction
A systematic approach of preprocessing and exploratory analysis of the dataset using Python and SQL integration via psycopg2 for PostgreSQL was the first step to creating useful visualisations and further analysis.
# Import the dataset to a dataframe
filename= 'data/dataset.csv'
df = pd.read_csv('data/dataset.csv')
pd.set_option("display.max_columns", None)
pd.set_option("display.max_rows", None)
# Check the amount of columns and rows
print(df.shape)
# >> (1552210, 44)
# Check the headers of columns
print(df.columns)
"""Index(['Unnamed: 0', 'Hour', 'HR', 'O2Sat', 'Temp', 'SBP', 'MAP', 'DBP',
'Resp', 'EtCO2', 'BaseExcess', 'HCO3', 'FiO2', 'pH', 'PaCO2', 'SaO2',
'AST', 'BUN', 'Alkalinephos', 'Calcium', 'Chloride', 'Creatinine',
'Bilirubin_direct', 'Glucose', 'Lactate', 'Magnesium', 'Phosphate',
'Potassium', 'Bilirubin_total', 'TroponinI', 'Hct', 'Hgb', 'PTT', 'WBC',
'Fibrinogen', 'Platelets', 'Age', 'Gender', 'Unit1', 'Unit2',
'HospAdmTime', 'ICULOS', 'SepsisLabel', 'Patient_ID'],
dtype='object')"""
# Cursory glance at snippet of table
print(df.head(15))
A significant size dataset with many null values in the first few rows printed. To get a clearer picture of how many values were missing and begin cleaning the dataset, I opted to import the table into a PostgreSQL and query from within Python using the library psycopg2.
Below is a function created to both query the Postgres database containing the table and export its query results into csv format if no existing csv of the same name is in the directory.
def export_query_to_csv(query, csv_file_path):
# Checks to see if file already present before committing function
if not os.path.exists(csv_file_path):
conn = psycopg2.connect(database="sepsis", user='postgres',
password='*******', host='localhost',
port='5432')
cur = conn.cursor()
# Generate the COPY command with the query
outputquery = "COPY ({0}) TO STDOUT WITH CSV HEADER".format(query)
with open(csv_file_path, 'w', newline='') as csv_file:
cur.copy_expert(outputquery, csv_file)
cur.close()
conn.close()
print("file saved successfully.")
else:
print("file already found.")
With our new function made, it is easier to perform the necessary queries and save the transformed results.
The first query was created to produce percentages of each columns missing values. Below is a snippet of part of the query.
query = """SELECT
ROUND(
(SUM(CASE WHEN Hour IS NULL THEN 1 ELSE 0 END) * 100.0 / COUNT(*)::float)::numeric, 2
) AS Hour_missing_percentage,
ROUND(
(SUM(CASE WHEN HR IS NULL THEN 1 ELSE 0 END) * 100.0 / COUNT(*)::float)::numeric, 2
) AS HR_missing_percentage,
......
FROM public.patient_data"""
csv_file_path = 'null_values.csv'
export_query_to_csv(query, csv_file_path)
To visualise query results, functions were created utilising Matplotlib and Seaborn. When called, this function would output a simple bar graph.
def query_visualization_bar(filename, xl, yl, title, columns_to_clean=None):
# Dictionary storing values from clean_column
data = {}
# opens csv and iterates through each row
with open(filename, 'r') as csv_file:
# using dictionary key-pairs to make data cleaning more streamlined
csv_reader = csv.DictReader(csv_file)
for row in csv_reader:
for column, value in row.items():
if column != "":
clean_column = column
if columns_to_clean:
for text_to_remove in columns_to_clean:
# Removing text in column header, specified when calling function.
clean_column = clean_column.replace(text_to_remove, '')
# Check if the value is an int
if value.isdigit():
data[clean_column] = int(value)
else:
# Try converting to float, use as-is if conversion fails
try:
data[clean_column] = float(value)
except ValueError:
data[clean_column] = value
# Extracts keys, values from cleaned dictionary
headers = list(data.keys())
values = list(data.values())
# Matplotlib bar plot
plt.figure(figsize=(12, 6))
plt.bar(headers, values, color='blue')
plt.xlabel(xl) # variable for x-axis label
plt.ylabel(yl) # variable for y-axis label
plt.title(title) # variable for title
plt.xticks(rotation=45, ha='right')
plt.tight_layout()
plt.show()
Here are the results from the previous query inputted into our bar graph function.
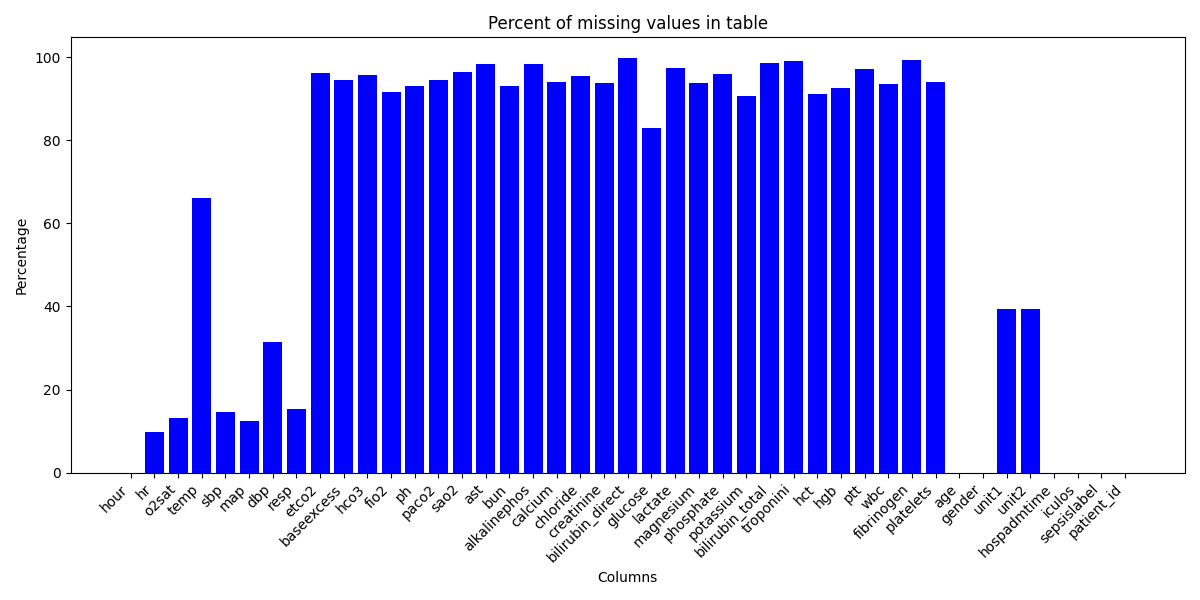 You can see that there are MANY missing values in our dataset, particularly those that are laboratory tested.
This makes sense because each row is a recording of a patients various health parameters at hourly intervals. Our more densely populated data relates to vital signs like heartrate (HR) and temperature (temp) as these are generally measured hourly for in-patients.
You can see that there are MANY missing values in our dataset, particularly those that are laboratory tested.
This makes sense because each row is a recording of a patients various health parameters at hourly intervals. Our more densely populated data relates to vital signs like heartrate (HR) and temperature (temp) as these are generally measured hourly for in-patients.
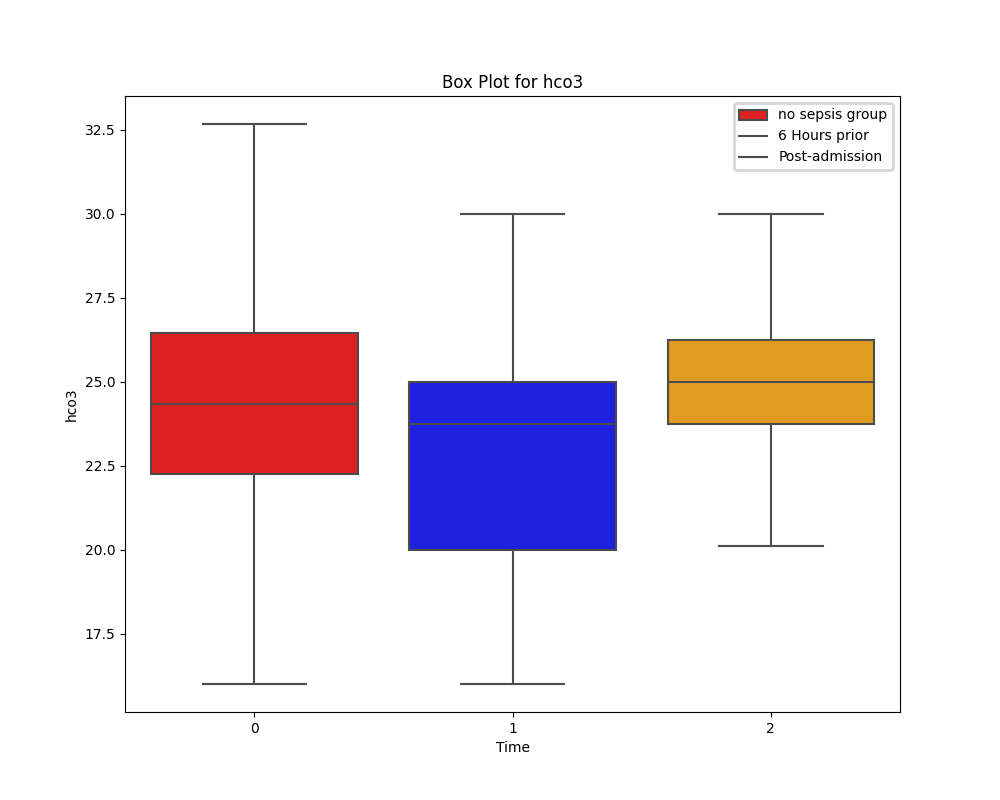
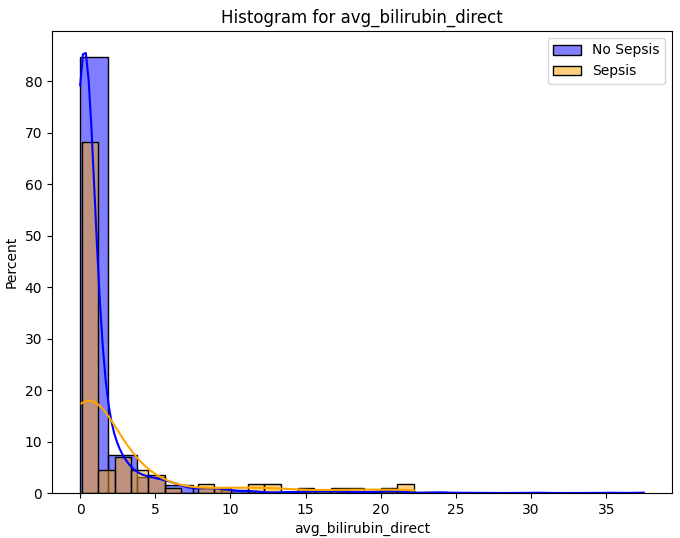
While tempting, it's important that we don't rush in and drop all of the columns with many missing values. Instead, appreciating the significance of these laboratory test and cleaning the data only as necessary appeared the best approach.

 You can see that there are MANY missing values in our dataset, particularly those that are laboratory tested.
This makes sense because each row is a recording of a patients various health parameters at hourly intervals. Our more densely populated data relates to vital signs like heartrate (HR) and temperature (temp) as these are generally measured hourly for in-patients.
You can see that there are MANY missing values in our dataset, particularly those that are laboratory tested.
This makes sense because each row is a recording of a patients various health parameters at hourly intervals. Our more densely populated data relates to vital signs like heartrate (HR) and temperature (temp) as these are generally measured hourly for in-patients.
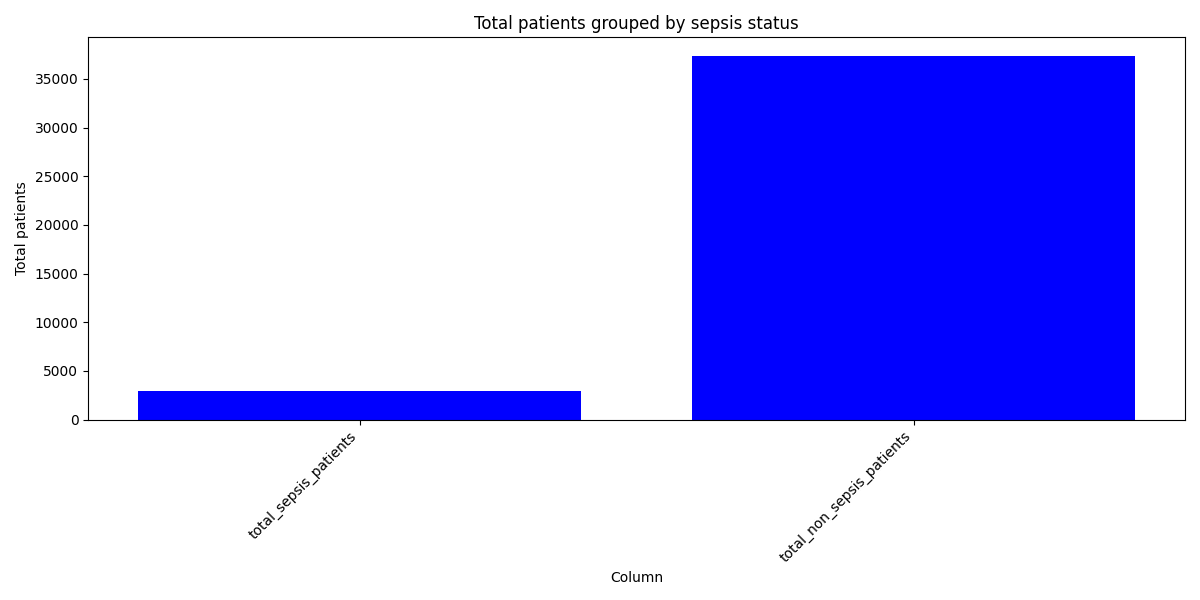
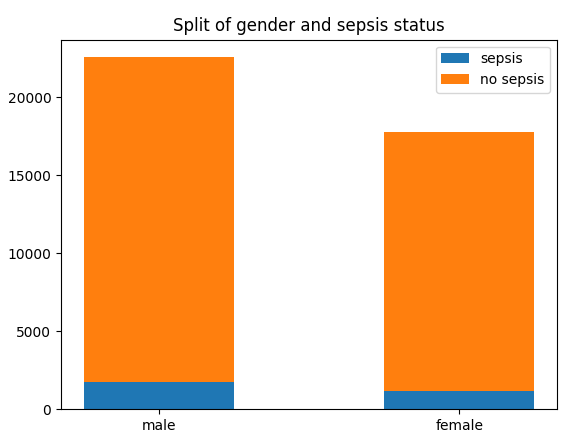
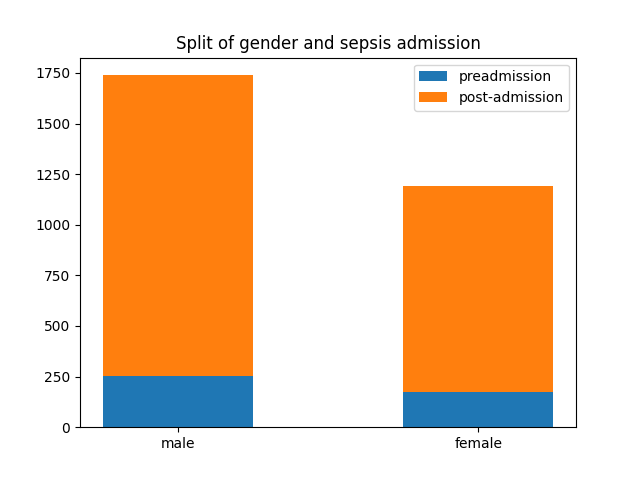
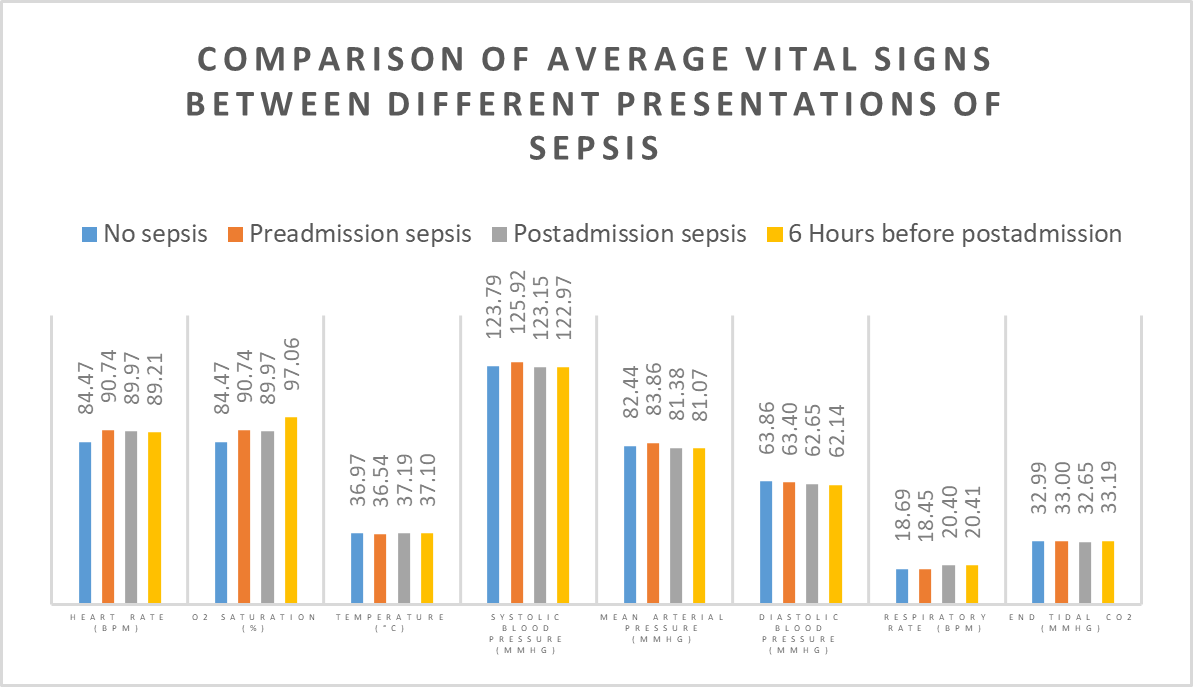
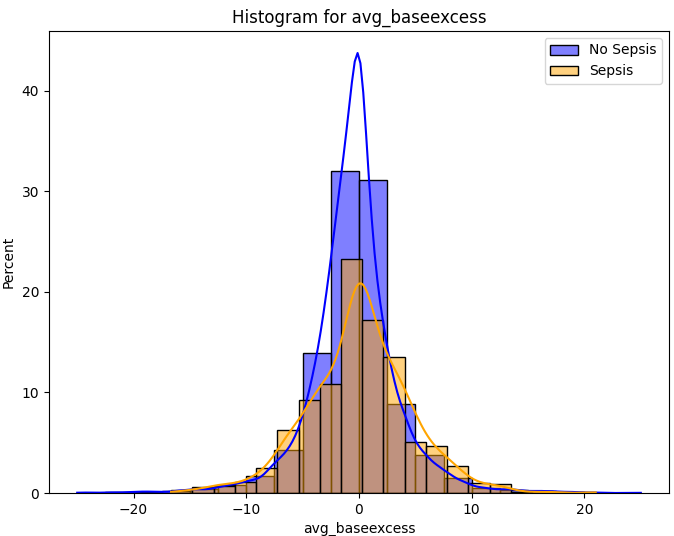
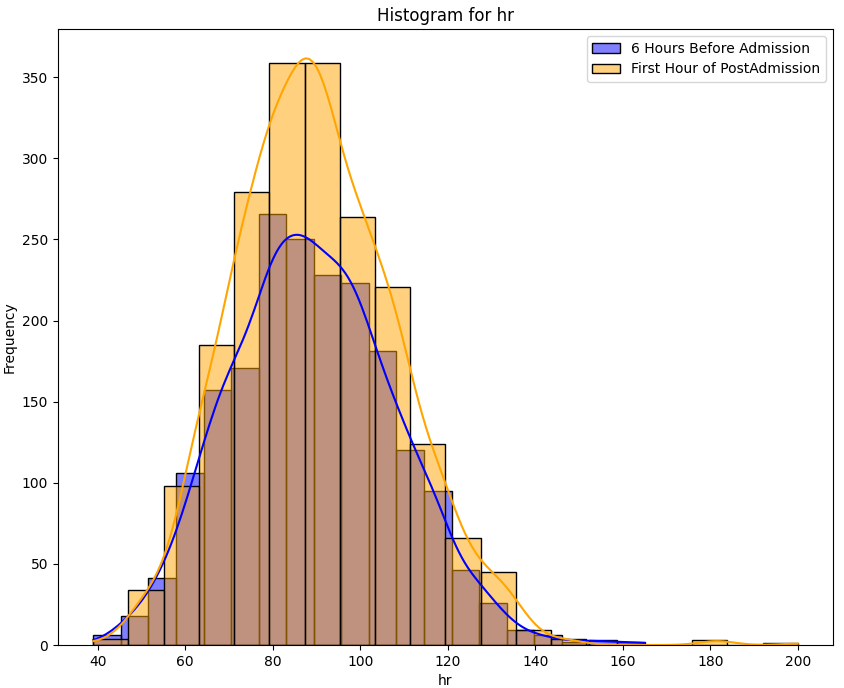
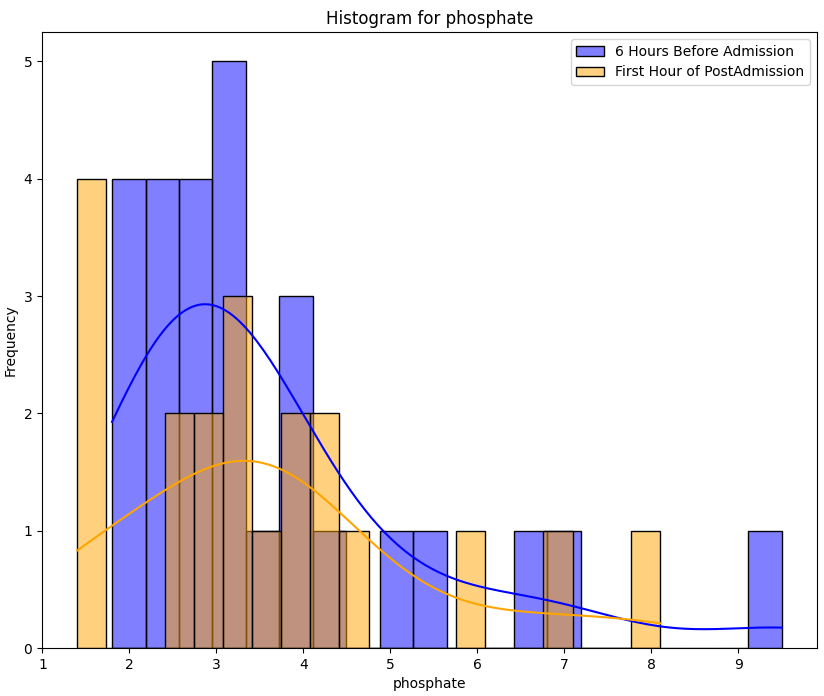
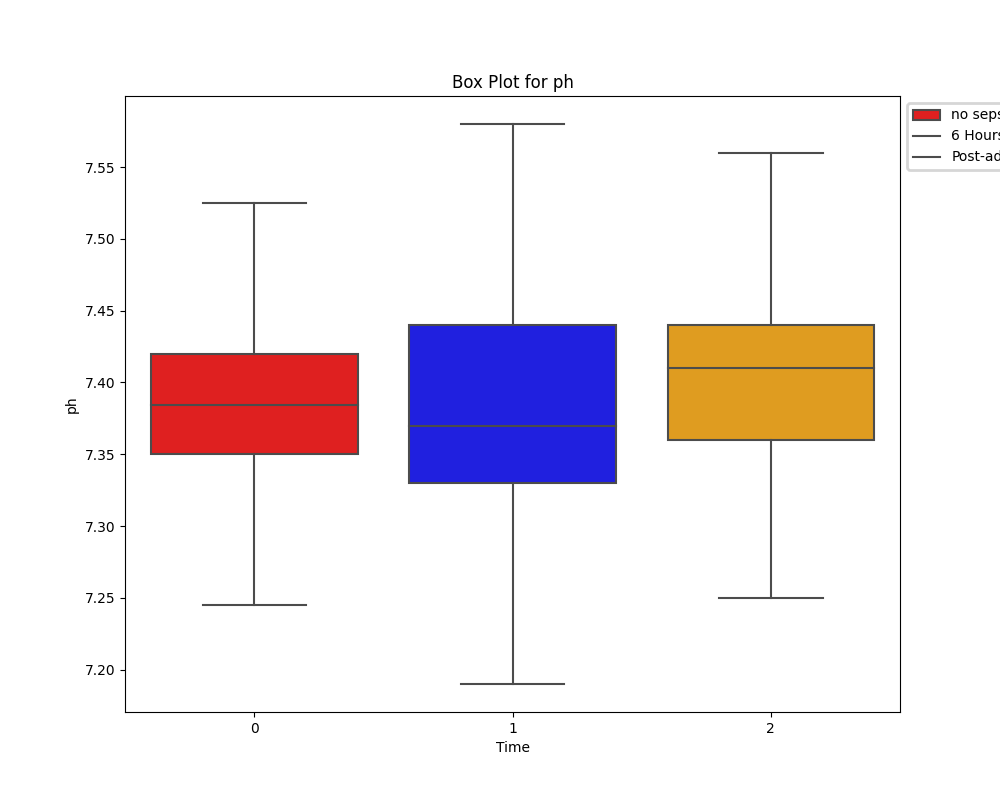
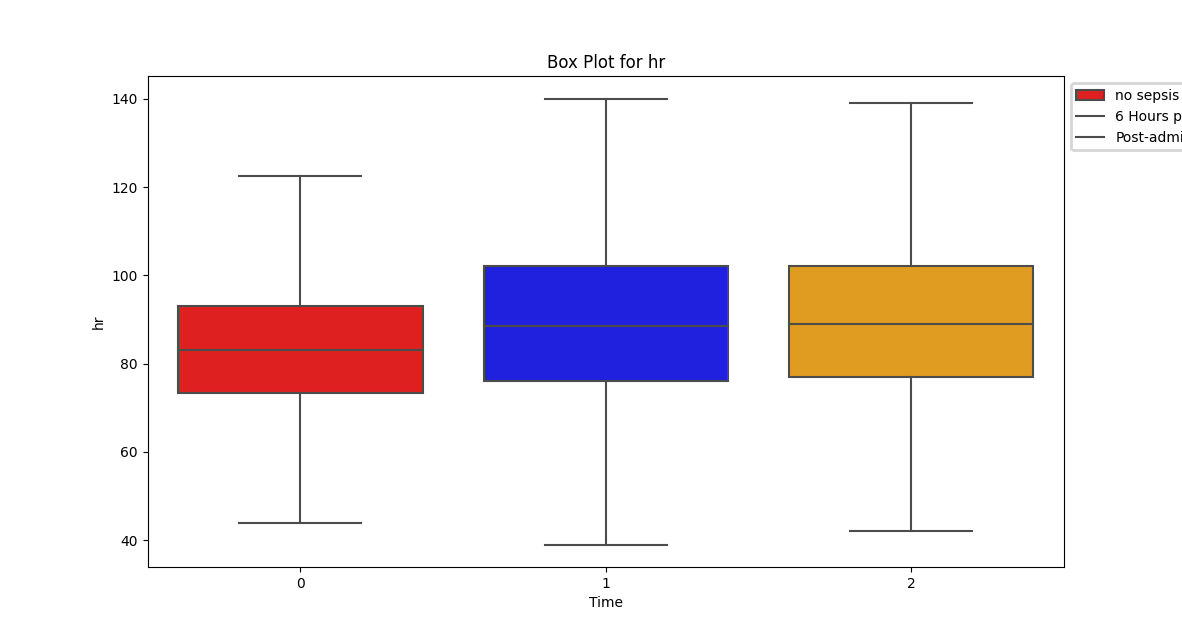 Despite a significant difference between means being found in our Wilcoxon Signed-Rank test, visually the distribution appears identical when looking at patients HR
six hours before confirming sepsis and at time of diagnosis. When compared to our larger population without sepsis, we can see that the distribution of pre-sepsis patients sits at a slightly elevated HR.
Despite a significant difference between means being found in our Wilcoxon Signed-Rank test, visually the distribution appears identical when looking at patients HR
six hours before confirming sepsis and at time of diagnosis. When compared to our larger population without sepsis, we can see that the distribution of pre-sepsis patients sits at a slightly elevated HR.
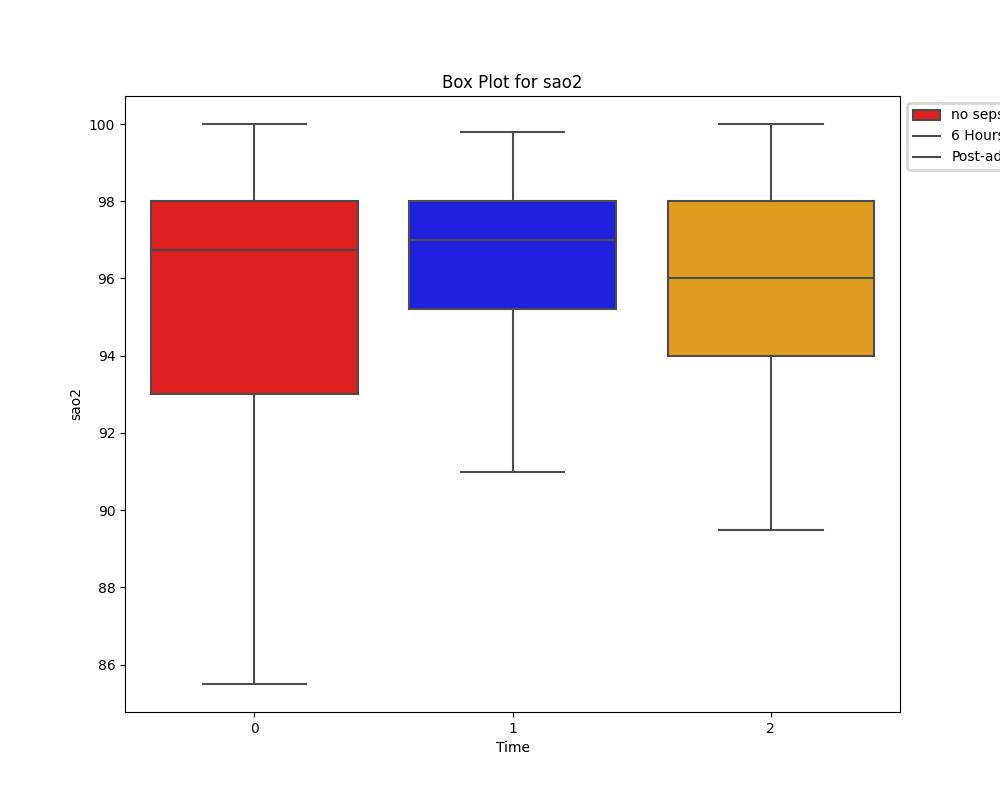
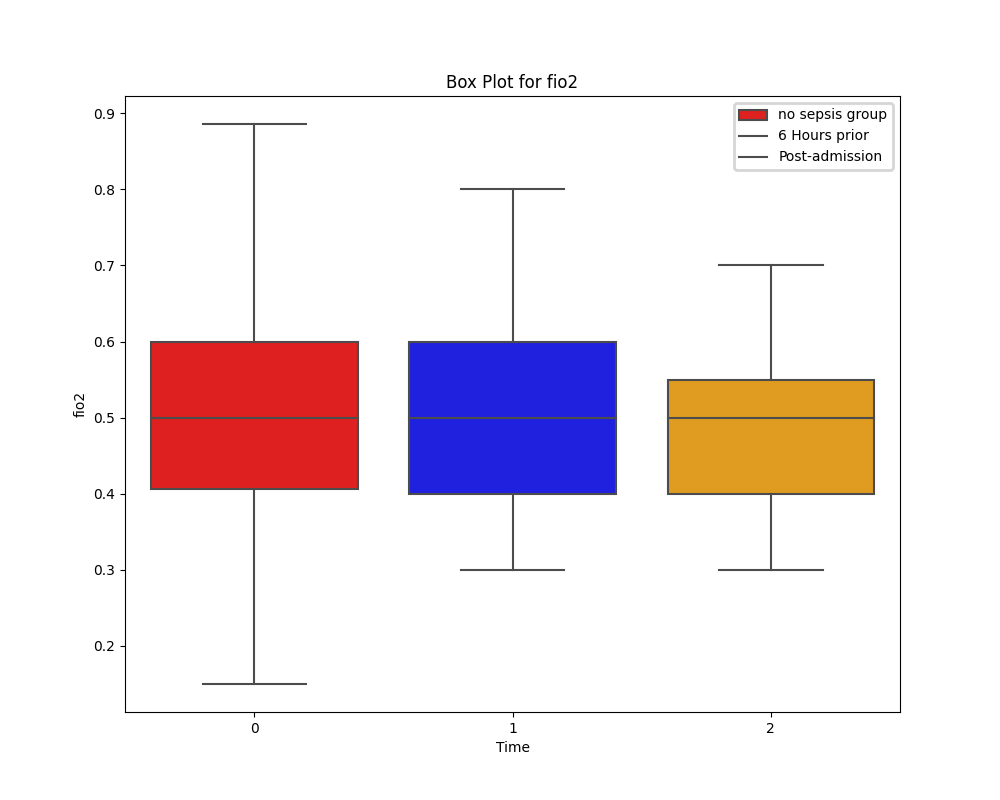
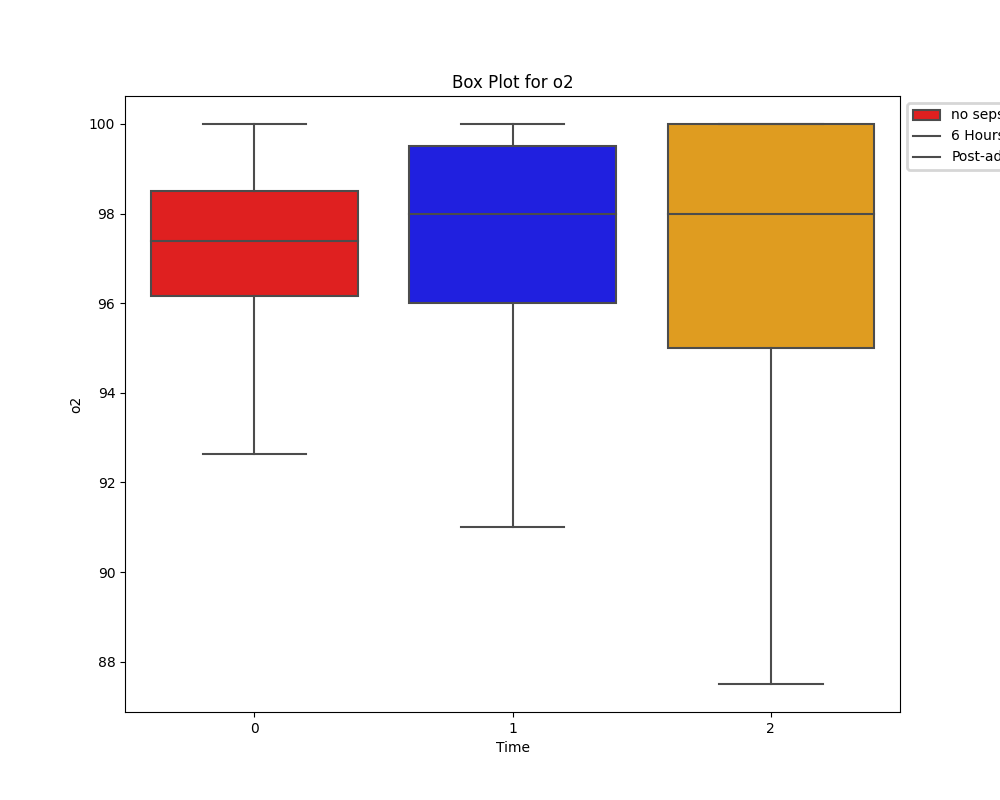 Unlike the distribution of HR, oxygen saturation does show a difference between the two groups. However, due to limitations of knowledge on respiratory therapies and any assitive ventialation strategies in place for these patients, its impossible to draw conclusions.
Unlike the distribution of HR, oxygen saturation does show a difference between the two groups. However, due to limitations of knowledge on respiratory therapies and any assitive ventialation strategies in place for these patients, its impossible to draw conclusions.